Funded Projects
2020 Projects
Interrogating Siglec Immune Checkpoints in Prostate Cancer
Postdoctoral Scholars:
Dr. Jessica Stark, Bertozzi Lab
Dr. Ru Wen, Brooks Lab
Dr. Scott Lovel, Bogyo Lab
Mentors:
Prof. Carolyn Bertozzi, Chemistry
Prof. James Brooks, Urology
Prof. Matthew Bogyo, Pathology and Microbiology and Immunology
Project Summary:
Despite the remarkable benefits of cancer immunotherapies observed in select cases, these molecules currently play a limited therapeutic role in prostate cancer. Thus, there is an urgent unmet need to identify additional immune checkpoints that drive disease progression. Emerging evidence suggests that upregulation of cell-surface glycoproteins modified with the sialic acid monosaccharide allows tumors to engage inhibitory glycan-binding receptors called Siglecs on immune cells. The objective of this work is to identify Siglec immune checkpoints that contribute to disease progression in prostate cancer as novel targets for cancer immunotherapies. Specifically, we will identify Siglecs that contribute to disease progression by comparing their expression to associated long-term clinical outcomes using human prostate cancer tissue microarrays (TMAs). Next, we will use an unbiased immunoprecipitation-mass spectrometry (IP-MS) approach to identify Siglec ligands expressed in prostate cancer cell lines and validate their roles in immune evasion in vitro. Finally, we will develop a glycopeptide array screening platform to determine epitopes bound by Siglecs in cancer, with an initial focus on Siglec-9. Within one year, this work will result in identification and initial molecular characterization of Siglec immune checkpoints in prostate cancer, laying the foundation for development of new checkpoint blockade immunotherapies.
Dissecting the Unique Invasion Machinery of the Malaria Parasite
Postdoctoral Scholars:
Dr. Marilou Tetard, Egan Lab
Dr. Stella Y. Sun, Chiu Lab
Dr. Li-av Segev Zarko, Boothroyd Lab
Mentors:
Prof. Elizabeth Egan, Pediatrics - Infectious Diseases
Prof. Wah Chiu, Bioegineering, Microbiology & Immunology, and Photon Science
Prof. John Boothroyd, Microbiology & Immunology
Project Summary:
he phylum of apicomplexan parasites includes several of the most prevalent and important human pathogens, causing malaria (Plasmodium), fatal diarrhea in children (Cryptosporidium) and severe neurological disease in the developing fetus and those who are immunocompromised (Toxoplasma). These obligatory intracellular, eukaryotic parasites enter a host cell by deploying a remarkable machine at their anterior end known as the apical complex (AC), for which the phylum is named. The AC is entirely specific to and highly conserved among the ~6000 species in the phylum, making it a rich target for development of novel drugs with broad specificity, a long-term goal of the work proposed here. To pursue this goal, we first need to understand how this multi-component machine coordinates invasion, an effort that until recently was not possible due to a lack of tools necessary for resolving the structure of such an apparatus in its native biological context. Cryo-electron microscopy (cryo-EM) has proven successful for delineating structures of single molecules at close to atomic resolution outside of their native environment. The challenge now is to overcome technical limitations to understand the structures of dynamic biological complexes in the eukaryotic cell, under different physiological or pathological states. The specific goal of the collaboration described here is, for the first time, to implement cryo-electron tomography (cryo-ET) to determine the precise spatial organization of the different molecular components of the AC in Plasmodium falciparum and P. knowlesi parasites, ultimately aiming for near-nanometer structural determination of the components in their native environment. We hypothesize that comparing such information with unpublished, parallel work accomplished by two of us (Segev-Zarko and Sun) in the better studied Toxoplasma will provide mechanistic insight into how members of this phylum accomplish the remarkable feat of invading human cells, as well as genus-specific solutions to the different challenges represented by the red blood cell, in the case of Plasmodium, and neurons and cardiomyocytes in the case of Toxoplasma.
2019 Projects
Charge Altering Releasable Transporters (CARTs) as a tunable platform for delivery of nucleic acids to the lung: new formulation for a novel universal therapeutic for influenza A virus (IAV)
Postdoctoral Scholars:
Dr. Timothy Blake, Waymouth Lab
Dr. David Paul Walton, Wender Lab
Dr. Rachel Hagey Saluti, Glenn Lab
Mentors:
Prof. Jeffrey Glenn, Medicine - Gastroenterology and Hepatology, Microbiology & Immunology
Prof. Robert Waymouth, Chemistry
Prof. Paul Wender, Chemistry, Chemical and Systems Biology
Project Summary:

Emerging DNA and RNA technologies have the potential to revolutionize medicine and life science research. Clinical progress of such therapeutics, however, is currently stifled by a lack of safe and effective delivery. Pulmonary drug delivery, in particular, is an area of clinical importance that suffers from these difficulties and is further confounded by the complexity of the respiratory tract, which has evolved to keep inhaled particles out and limit cellular uptake. Charge Altering Releasable Transporters (CARTs) developed by the Wender-Waymouth laboratories encompass a platform technology that promises safe and effective delivery of nucleic acids, with the potential to propel DNA and RNA therapeutics through the clinic. CARTs already outperform commercial reagents and allow the targeting of more challenging organs and tissues. With this ChEM-H seed grant initiative, we seek to apply the CART platform technology to develop a first-of-its-kind formulation for delivery of a novel nucleic acid inhibitor with broad-spectrum activity against influenza A virus developed by the Glenn laboratory, specially optimized for the lung. This proof-of-principle work has a path to the clinic as a nebulized universal flu therapeutic possessing a high barrier to antiviral resistance, and serves to pave the way for similar nucleic acid delivery strategies in other disease models of clinical interest.
Development of Novel Molecular Imaging Agents for Visualization of Cytotoxic T-cells and evaluation of CAR-T cell therapy in preclinical models of glioblastoma
Postdoctoral Scholars:
Dr. Corinne Beinat, Gambhir Lab
Dr. Chirag Patel, Recht Lab
Mentors:
Prof. Sanjiv Sam Gambhir, Radiology
Prof. Lawrence Recht, Neurology and Neurological Sciences
Project Summary:

Glioblastoma is the most common and lethal type of primary brain cancer, with half of patients dying within 14-18 months of their diagnosis, and 5.6% of patients alive five years after their diagnosis. One promising approach for the treatment of glioblastoma is immunotherapy. Immunotherapy is a type of cancer treatment that boosts the body’s natural immune system to fight cancer. Immunotherapy involves natural or laboratory-manufactured agents to amplify or restore immune system function, which in turn helps our body’s immune system to recognize and destroy cancer cells. One of the key challenges in the successful treatment of glioblastoma with immunotherapy is the lack of appropriate methods to visualize and monitor the killing of cancer cells by the immune system. This proposal aims to overcome this challenge by developing a novel imaging agent to visualize and quantify perforin, a specific protein that immune cells use to kill cancer cells. This would allow clinicians and researchers to visualize and have real-time information of the killing of cancer cells by the immune system and make informed decisions about the effectiveness of immunotherapy for individual patients with glioblastoma.
2018 Projects
Size-based Isolation of Extracellular Vesicles from iPSC-cardiomyocyte for Diagnosis of Cardiomyopathies
Postdoctoral Scholars:
Dr. Mark Chandy, Wu Lab
Dr. Mehmet Ozen, Demirci Lab
Mentors:
Prof. Joseph Wu, Cardiovascular Medicine and Radiology
Prof. Utkan Demirci, Radiology
Project Summary:

Cardiomyopathy is a severe disease of the heart that leads to heart failure—the heart cannot keep up with the demands of the body and fluid builds up in the lung and body. If a cardiomyopathy is detected early, physicians can start medications and use devices to improve symptoms and prolong life. We propose to search for new biomarkers of heart failure by studying the vesicles secreted from the heart. To do this, we will generate heart tissue from patients using stem cells. We will then utilize an engineering approach to isolate microvesicles and compare their contents with patient blood and identify novel biomarkers to diagnose cardiomyopathy.
Investigating the Enteric Nervous System Metabolome
Postdoctoral Scholars:
Dr. Subhamoy Das, Kaltschmidt Lab
Dr. Vijaya Lakshmi Kanchustambham, Zare Lab
Mentors:
Prof. Julia Kaltschmidt, Neurosurgery
Prof. Richard Zare, Chemistry
Project Summary:

Digestion is an important physiological phenomenon facilitated by nerves, muscles, and other gut cells. Various gastrointestinal disorders cause disruption of the normal digestion process and many of these have a neuronal origin. The nervous system that resides within the gut wall is called the enteric nervous system (ENS), also referred as the “second brain”. Understanding the biochemical profile of the ENS is a long-standing open question. While the ENS field has relied heavily on immuno-labeling of peptides and proteins such as neurotransmitters and signaling molecules, studying the entire metabolic profile of the ENS presents a serious challenge that so far has never been met. Speed and consistency limit standard techniques to analyze metabolites over the length of the entire gut tube. Here we propose to use a new, high-throughput technology called DESI-MSI (Desorption ElectroSpray Ionization - Mass Spectrometry Imaging). DESI-MSI can be used to scan tissue micro-sections and acquire the entire metabolic profile of the native tissue via mass spectrometry. This metabolic profile includes neurotransmitters, lipids, small molecules, small peptides, and other metabolites. In this proposal, we aim to scan mouse tissue sections from each portion of the gut (from the beginning of the esophagus to the end of the colon) and create a spatial map of the metabolic signature of the ENS. In the future, this information will drive drug discovery and the development of therapeutics for gastrointestinal diseases.
Connectome-seq: Unbiased High-throughput Brain Connectome Mapping
Postdoctoral Scholars:
Dr. Alina Isakova, Quake Lab
Dr. Boxuan Zhao, Ting and Luo Labs
Mentors:
Prof. Stephen Quake, Bioengineering and Applied Physics
Prof. Alice Ting, Genetics and Biology
Prof. Liqun Luo, Biology
Project Summary:

The ability of the brain to control our behavior, feelings, and thoughts has long been of a great interest and fascination to scientists. Unveiling the complexity of the brain and particularly its intricate network of connections, controlling various physiological functions of the body, remains one of the biggest scientific challenges of the 21st century. This, however, proved to be a non-trivial task, mainly due to limitations of the current technologies that are unable to capture the brain connectome at the full scale. Here, we set out to address this challenge and create a state of the art technology capable to map extensive networks of neural connections across proximal and distal regions of the brain. Combining the most recent discoveries in the fields of molecular biology and engineering, we will create a trans-synaptic labeling system aimed to effectively tag all synapses in the brain with unique nucleic acid barcodes. Once tagged, the synapses can then be read out by high-throughput sequencing, revealing the identity of neurons involved in the interaction. Profiling a vast number of synapses, we expect to fully map the interactions between various types of neurons and thus reconstruct a global map of connectome linking different brain regions.
Monitoring Tumor Heterogeneity and Drug Response in Barcoded Patient Derived Organoids Using Multi-omics Single Cell Approach
Postdoctoral Scholars:
Dr. Kasper Karlsson, Curtis Lab
Dr. Amber Smith, Kuo Lab
Dr. Akshay Balsubramani, Kundaje Lab
Kathryn Yost, Chang Lab
Mentors:
Prof. Christina Curtis, Medicine - Oncology and Genetics
Prof. Calvin Kuo, Medicine - Hematology
Prof. Anshul Kundaje, Genetics and Computer Science
Prof. Howard Chang, Dermatology and Genetics
Project Summary:

Pancreatic Ductal Adenocarcinoma (PDAC) is an insidious and deadly malignancy, with the worst 1-year and 5-year survival rates of all cancer types. Poor survival is in part due to our incomplete understanding of tumor heterogeneity, and how tumor sub-clones influence drug response. To increase our understanding of PDAC heterogeneity we will derive organoids from resections of PDAC tumors and make cells unique by integrating a random DNA barcode into the genome of each cell. Since daughter cells inherit the integrated barcode, cells with the same barcode can be defined as a tumor sub-clone. Barcoded cells will be expanded, and drug treated with standard of care drugs for PDAC to identify sub-clone specific response. Single cell sequencing in combination with barcode sequencing will be used to elucidate transcriptome, regulome and genome composition of resistant sub-clones, in order to identify novel biomarkers for sub-clone resistance.
2017 Projects
Wireless Monitoring of Blood Flow Using Biodegradable and Flexible Pressure Sensors
Postdoctoral Scholars:
Dr. Levent Beker, Bao Lab
Dr. Yukitoshi Kaizawa, Fox and Chang Labs
Mentors:
Prof. Zhenan Baro, Chemical Engineering
Prof. Paige Fox, Plastic Surgery
Prof. James Chang, Plastic Surgery
Project Summary:

An increasing number of patients need surgical interventions that require small vessel anastomoses, which are critical to surgical success. However, current methods of monitoring blood flow post-anastomosis require trained medical staff and/or specialized equipment. Because of the lack of continuous monitoring, often times, graft failure is not noted until the opportunity to save the graft has passed. We propose a wireless blood flow measurement method that does not require trained medical staff or expensive equipment. The planned method utilizes a simple, flexible, and biodegradable sensor that will allow surgeons to monitor blood flow across vascular anastomoses continuously after surgery which would lead to less morbidity. Additionally, it could eliminate monitoring tests, decrease frequent subjective human evaluations, and limit time consuming clinic visits. In this study, a pressure sensor to detect expansion of an artery will be fabricated, and in-vivo tests will be conducted to verify the operation of the sensor. The sensor will be designed to send data wirelessly via inductive coupling and operate without battery due to its passive structure. Outcomes of this research can also be expanded to other medical applications such as treatment of brain aneurysms.
Publications:
Biodegradable and flexible arterial-pulse sensor for the wireless monitoring of blood flow. C.M. Boutry, L. Beker, Y. Kaizawa, C. Vassos, H. Tran, A.C. Hinckley, R. Pfattner, S. Niu, J. Li, J. Claverie, Z. Wang, J. Chang, P.M. Fox, Z. Bao. Nature Biomedical Engineering. 2019, 3, 47-57.
Identifying Medically and Therapeutically Relevant GPCR Genetic Variants Based on 500,000 Patients
Postdoctoral Scholars:
Dr. Christopher DeBoever, Bustamante Lab
Dr. A.J. Venkatakrishnan, Dror and Kobilka Labs
Mentors:
Prof. Carlos Bustamante, Biomedical Data Science and Genetics
Prof. Ron Dror, Computer Science
Prof. Brian Kobilka, Molecular & Cellular Physiology
Project Summary:

About three in five American adults take prescription drugs every month and many experience adverse drug reactions or reduced treatment efficacy. Nearly one-third of all FDA-approved drugs target ~100 G protein-coupled receptors (GPCRs), a family of proteins embedded in the cell membrane. Inherited genetic variants are known to affect disease risk and drug response, yet little is known about variants in GPCRs. Which genetic variants in GPCRs are associated with disease and diversity in drug response? What is the mechanism through which genetic variants in GPCRs affect health and drug response? We propose to answer these questions by identifying genetic variants that are associated with disease or adverse drug responses using data from 500,000 patients and studying the effect of these variants using protein structure-based modeling. Our cross-discipline approach will identify high-confidence genetic variants in GPCRs that can be used to understand the limitations of current drugs, develop new drugs, and ultimately improve patient care.
Examining the Role of Neuron-Glia Signaling in Pain Perception
Postdoctoral Scholars:
Dr. Erin Elizabeth Gray, Du Bois Lab
Dr. Husniye Kantarci, Zuchero Lab
Mentors:
Prof. Justin Du Bois, Chemistry
Prof. Brad Zuchero, Neurosurgery
Project Summary:
Chronic pain affects millions of people worldwide, but many existing therapeutic treatments exhibit limited efficacy or serious side effects. To improve the treatment of pain, a deeper understanding of the complex mechanisms that underlie nociception is necessary. The electrical impulses generated in response to noxious stimuli are conducted in excitable cells by voltage-gated sodium channels (NaVs). Although the excitability of neurons is thought to be controlled intrinsically, we have recently discovered that sensory neurons require signaling from myelinating glia to maintain excitability. This proposal seeks to elucidate the underlying mechanism(s) by which glia-derived molecules modulate NaV activity. Our studies will capitalize on methods developed in the Du Bois and Zuchero groups, including novel molecular probes for labeling and imaging NaVs and state-of-the-art techniques for purifying neurons and glia. Further mechanistic insight into this regulatory pathway will inform our understanding of neuronal excitability and may lead to the identification of new pharmaceutical targets for the treatment of pain.
Identification of Biased Positive Allosteric Modulators of Cannabinoid Receptor Type 1 for the Treatment of Neuropathic Pain
Postdoctoral Scholars:
Dr. Kaavya Krishna Kumar, Kobilka Lab
Dr. Angel Resendez, Molhotra Lab
Mentors:
Prof. Brian Kobilka, Molecular & Cellular Physiology
Prof. Sanjay Malhotra, Radiation Oncology and Radiology
Project Summary:

Neuropathic pain is a debilitating form of chronic pain caused by damage or disease to the nervous system. It is estimated to affect 1 in every 10 adults over the age of 30 and exacts a significant healthcare cost, rehabilitation and lost worker productivity. In addition, neuropathic pain places an enormous financial and emotional burden on patients and their families. Neuropathic pain remains a significant clinical problem because it responds poorly to the available therapies. Hence there is a need to identify novel analgesic targets for drug development. Evidence for the use of Cannabis sativa as a treatment for pain can be traced back to the beginnings of recorded history. It acts through the stimulation of the cannabinoid type 1 receptor (CB1R), one of the most abundantly expressed G protein-coupled receptors (GPCRs) in the central nervous system. Targeting CB1R has huge therapeutic potential in the treatment of neuropathic pain, however, psychotropic side effects preclude the widespread use of cannabinoid drugs. This proposal aims to identify molecules that would allow CB1R to preferentially signal via pathways that cause pain relief without the side effects. The outcomes of this proposal will have profound implications in the treatment and management of neuropathic pain.
2016 Projects
CRISPRouting Neural Development in a Zebrafish Model
Postdoctoral Scholars:
Dr. P.C. Dave P. Dingal, Qi Lab
Dr. Karen Mruk, Chen Lab
Mentors:
Prof. James Chen, Chemical and Systems Biology & Developmental Biology
Prof. Stanley Qi, Bioengineering & Chemical and Systems Biology
Project Summary:
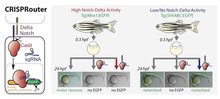
The Notch-Delta receptor signaling – triggered via direct cell-to-cell contact – plays a major role in embryonic development. In the central nervous system (CNS), Notch-Delta signaling controls the cell numbers and fate choice of specific neurons. However, both loss-of-function and gain-of-function mutants in this pathway lead to developmental defects. Conditional expression systems are perhaps our best tools, for understanding the role of Notch-Delta signaling in regulating neuronal cell fate.
This proposal aims to apply molecular engineering and synthetic biology approaches to engineer the Notch receptor and the CRISPR-Cas9 system for conditional knockout and overexpression studies in zebrafish. The zebrafish is a powerful animal model for these studies as they are completely transparent permitting real-time visualization of development. We have recently developed a class of human Notch-Cas9 chimeric receptors (CRISPRouters) that binds to extracellular Delta and releases receptor-tethered Cas9, a DNA-binding enzyme that we can program to target specific genes. Applying to zebrafish models, we will introduce CRISPRouters to perturb and examine the dynamic role of the Notch-Delta pathway during nervous system development. The modular CRISPRouter toolkit will allow rewiring of cell-cell signaling and cell fate, providing a novel approach to developmental studies.
Genome-Wide Screen for Regulators of Adipogenesis
Postdoctoral Scholars:
Dr. Kyuho Han, Bassik Lab
Dr. Keren Hilgendorf, Jackson Lab
Mentors:
Prof. Michael Bassik, Genetics
Prof. Peter Jackson, Microbiology and Immunology & Baxter Laboratories
Project Summary:

Obesity and co-morbidities like diabetes are a major unmet medical need, and there are few effective therapies targeting obesity or adipocyte pathology. To better understand the pathogenesis of unhealthy fat, we are interested in understanding how fat differentiation is initiated and regulated. Specifically, we will perform a genome-wide knockout screen using a fat progenitor cell line that robustly differentiates in vitro into lipid-laden fat cells upon exposure to differentiation factors. This screen will identify both known and novel genes required for fat differentiation. We are particularly interested in identifying genes related to primary cilia function. The cilium is a hair-like cellular protrusion that senses biochemical signals. In fat progenitor cells, it senses insulin and new factors we have identified. How primary cilium signaling is integrated to initiate fat differentiation remains unclear. In parallel, we will investigate the epigenetic changes underlying differentiation. In response to differentiation cues, fat progenitor cells undergo two rounds of cell division, which cue expression of differentiation-specific proteins. The regulation of these divisions and the specific transition that changes cell fate is also not known. Understanding the molecular factors regulating fat differentiation may uncover novel targets that lessen the burden of obesity.
LRET Based Single Molecule ISH for Genomic Imaging in Tissues
Postdoctoral Scholars:
Dr. Nicholas Juul, Desai Lab
Dr. Monica Nagendran, Harbury Lab
Mentors:
Prof. Tushar Desai, Medicine - Pulmonary & Critical Care Medicine
Prof. Pehr Harbury, Biochemistry
Project Summary:

We are developing a rapid, scalable, automated, and easy-to-use technology for measuring global gene expression at the single-cell level in intact tissues. This technology, which we call ‘Genomic Imaging,’ will rapidly replace single-cell RNA-sequencing and conventional in situ hybridization techniques, because it merges the deep gene coverage of the former with the spatial resolution of the latter. The technology is based on directly marking transcripts in fixed tissues, which avoids the reverse transcription and amplification steps that introduce bias in sequencing workflows. Genomic Imaging is compatible with concurrent antibody staining, and works robustly on formalin-fixed, paraffin-embedded (FFPE) tissue. We predict that the generation of “molecular histology” atlas's will become a widely adopted research assay, and the standard for molecular profiling across all medical disciplines.
Immunostimulatory Synthetic Glycopeptides Targeting a Novel Pathway for Cancer Immunotherapy
Postdoctoral Scholars:
Dr. Justin Kenkel, Engleman Lab
Dr. Jessica Kramer, Bertozzi Lab
Mentors:
Prof. Edgar Engleman, Pathology & Medicine - Immunology & Rheumatology
Prof. Carolyn Bertozzi, Chemistry
Project Summary:
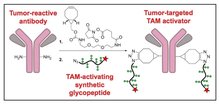
Cancer immunotherapies show great promise in the clinic, but only a fraction of patients with certain cancers responds to existing immunotherapies. We recently discovered a new way to stimulate tumor immunity by targeting a pattern recognition receptor expressed by tumor-associated macrophages (TAMs). We found that triggering this receptor with microbial extracts reprograms TAMs into immunostimulatory cells that induce antitumor responses. The natural ligands for this receptor inhibit tumor progression as single agents and synergize with conventional therapies (e.g. chemotherapy) and other immunotherapies (e.g. checkpoint blockers) to induce tumor regression. Encouraged by these results, we decided to use a novel synthetic approach to prepare chemically defined, glycopeptide-based receptor ligands. With this platform, we can easily modify glycopeptide structure and function to generate strong TAM stimuli that can be targeted to tumor tissues and combined with other immunostimulatory molecules. We have already synthesized a series of glycopeptides, and our preliminary studies fully support this strategy. We are now developing glycopeptide agonists optimized for in vivo activity and functionalized with tumor-targeting properties. We expect this exciting project combining chemistry and tumor immunology to lead to the development of adaptable TAM activators that will form the foundation of a new class of cancer immunotherapeutics.
A Novel Proteolytic Profiling Assay Based on Spectrally Encoded Beads to Guide the Design of Selective Chemical Probes for Bacterial Pathogens
Postdoctoral Scholars:
Dr. Chrstian Lentz, Bogyo Lab
Dr. Huy Nguyen, Fordyce Lab
Mentors:
Prof. Matthew Bogyo, Pathology & Microbiology and Immunology
Prof. Polly Fordyce, Genetics & Bioengeering
Project Summary:

Chemical probes that visualize the activity of members of the protease enzyme family are useful tools for biomedical research and in vivo imaging. One of our long-term goals is to develop probes that would specifically recognize single proteases of bacterial pathogens and could be used for non-invasive in vivo imaging of infection. Protease-directed probes are usually based on amino acid sequences that mimic the natural substrates of a protease. However, probes based on the limited combinations of the 20 naturally occurring amino acids may lack specifity. Introducing chemical diversity by incorporating some of the >100 available non-natural amino acids may enhance these probes tremendously, but there are currently no flexible, cost-effective, high-throughput screening systems available to identify which non-natural amino acids individual proteases prefer.
2015 Projects
The Effect of Pediatric Specific Hypertrophic Cardiomyopathy Mutations on the Biomechanics of beta-Cardiac Myosin at the Molecular and Cellular Level
Postdoctoral Scholars:
Dr. Arjun Adhikari, Spudich Lab
Dr. Kristina Bezold, Bernstein Lab
Dr. Alexandre Ribeiro, Pruitt Lab
Mentors:
Prof. James Spudich, Biochemistry
Prof. Daniel Bernstein, Pediatrics - Cardiology
Prof. Beth Pruitt, Mechanical Engineering
Project Summary:
Hypertrophic Cardiomyopathy (HCM) is a genetic disease that affects 1 in 500 people. In infants it is particularly severe and is the leading cause of sudden cardiac death in pediatric populations. A high percentage of HCM is attributed to mutations in ß-cardiac myosin, the protein responsible for heart contraction. This study explores how pediatric specific HCM mutations can alter the biochemical and biomechanical properties of ß-cardiac myosin at a molecular and cellular level. At a molecular level, this study aims to understand how HCM mutations affect the biomechanics of purified human ß-cardiac myosin. At a cellular level, cardiomyocytes derived from human induced pluripotent stem cells (iPSC) engineered to express each mutation will be used to study how HCM affects the power output and signaling pathways of these cells. The current standard care for patients with HCM involves beta-blockers and ACE inhibitors, which treat the symptoms, but not the disease. The results from this study will increase the understanding of how genetic mutations that cause HCM lead to the presentation of the disease, and will uncover potential targets for therapeutic design to treat the cause of the disease instead of the symptoms.
This project brings together three postdoctoral researchers to form an interdisciplinary team spanning the wide range of skill sets necessary to study this important problem in cardiac disease. Dr. Adhikari is an expert in biophysics and cell biomechanics and has solid knowledge of cell and molecular biology. During his doctoral work with Dr. Dunn (Stanford Chemical Engineering), he gained deep understanding of how mechanical forces can influence biological function at a molecular and cellular level. He is currently a postdoctoral researcher in Dr. James Spudich’s lab in the Biochemistry Department. Dr. Bezold is an expert in muscle biology and biomechanics and has significant experience in stem cell biology. She gained in depth knowledge of muscle biology while working in the lab of Dr. Samantha Harris (UC Davis). Currently, she is a postdoctoral fellow in the lab of Dr. Daniel Bernstein in Pediatrics/Cardiology, where she has led the effort to generate iPSC lines with HCM mutations using the CRISPR/Cas9 system. Dr. Ribeiro is a highly skilled mechanical engineer and has considerable experience in cell biology. During his graduate work, under the guidance of Dr. Kris Dahl (Chemical Engineer, Carnegie Mellon University), he studied nuclear mechanics during cell migration. During his postdoctoral work in the lab of Dr. Beth Pruitt in Mechanical Engineering, he has been instrumental in developing patterned platforms that lead to more mature phenotype of iPSC cardiomyocytes.
Bioorthogonal Secretome Labeling for the Study of Mammalian Aging and Rejuvenation
Postdoctoral Scholars:
Dr. Kyle Brewer, Wyss-Coray Lab
Dr. Justin Kim, Bertozzi Lab
Mentors:
Prof. Tony Wyss-Coray, Neurology & Neurological Sciences
Prof. Carolyn Bertozzi, Chemistry
Project Summary:
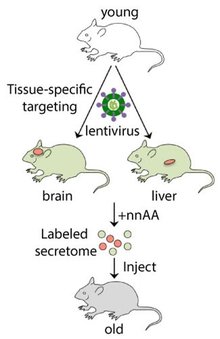
Aging is the largest risk factor for heart disease, cancer, stroke, dementia, and diabetes, as well as a major impediment to quality of life. Recently, protein factors present in young blood have been shown to rejuvenate the heart, brain, and muscle in aged mice. In addition, protein factors in old blood have been shown to impair cognitive function in mice. To find how these proteins can best be exploited to benefit human health, tissues in mice will be examined to determine where these factors are produced, as well as where they localize. To achieve this goal, recent developments in chemical biology will be used to incorporate non-natural amino acids (nnAAs) into the mouse proteome to target secreted factors for bioorthogonal labeling. The genetic system to incorporate the nnAAs will be introduced specifically into the brain and liver of young mice via lentiviral delivery, and the proteins secreted from these regions will be collected in plasma. After injecting the labeled, young plasma into unlabeled, old mice, whether these factors can cross the blood-brain barrier and where these factors distribute in the brain will be determined. This system can then be applied more widely to find other areas these proteins have specific effects to rejuvenate tissues.
This project leverages the complementary expertise of two postdoctoral researchers with knowledge of protein and synthetic chemistry to address an important question in human biology. Dr. Brewer has a background in biophysics including protein structure, protein dynamics, and protein-protein interactions and has extensive experience using novel compounds and biochemistry to produce labeled proteins for biophysical and biological purposes. Since joining the Wyss-Coray Lab as a postdoctoral researcher, he has been developing novel methods to study the mouse secretome, placing an emphasis on examining natively and in vitro the aging and rejuvenation factors being discovered in the lab. Dr. Kim’s graduate work was performed in the field of complex natural product total synthesis, and he has expertise in non-natural amino acid incorporation and bioorthogonal labeling. His postdoctoral research in the Bertozzi Lab involves the use of non-natural amino acids in conjunction with bioorthogonal chemistry to study the mechanism by which secreted virulence factors mediate the pathogenesis of tuberculosis in a Mycobacterium marinum-zebrafish model system.
A Chemical-Genetic Investigation of Non-Apoptotic Cell Death Mechanisms
Postdoctoral Scholars:
Dr. Jennifer Yinuo Cao, Dixon Lab
Dr. Cole Dovey, Carette Lab
Mentors:
Prof. Scott Dixon, Biology
Prof. Jan Carette, Microbiology & Immunology
Project Summary:

Regulated cell death enables the elimination of old, damaged or unwanted cells from the body. This process is essential to maintain homeostasis and prevent disease. A central aim of the Carette and Dixon laboratories is to better understand the structure and function of different cell death pathways which will ultimately allow substantial improvements in human health. The goal of this proposal is to better understand the operation of two recently described non-apoptotic cell death pathways, termed necroptosis and ferroptosis. The regulation of these pathways at the genetic and biochemical levels remains largely a mystery. To address this problem, this proposal combines state-of-the-art methods in human genetics and chemical biology pioneered in the Carette and Dixon laboratories. Specifically, a powerful, high-throughput chemical screening approach will be applied to identify small molecule modulators of necroptosis. Concurrently, an innovative, genome-wide genetic screening platform will be employed to investigate the genetic architecture of ferroptosis. This research aims to clarify the regulation of non-apoptotic cell death pathways and identify new druggable targets that ultimately will enable more precise therapeutic control of cell death.
Dr. Cao completed her doctoral studies in the Frappier Lab at the University of Toronto, where she used proteomic and biochemical techniques to identify novel herpes virus-host protein interactions which revealed novel connections between herpes virus and reactive oxygen species (ROS) production. She is currently studying the regulation of ferroptosis under the mentorship of Dr. Scott Dixon and has helped develop a novel high-throughput, time-lapse imaging platform that can be used to examine the effects of various bioactive small molecules on cell death. Dr. Dovey has over a decade of experience in molecular pathogenesis research, with expertise in genetics and biochemistry. During his graduate training with Jeffery Cox at UCSF, he applied a biochemical and bacterial genetic approach toward understanding the regulation of virulence in Mycobacterium tuberculosis. As a postdoctoral scholar in the Carette Lab, he has expanded his field of expertise by investigating the cell biological and genetic mechanisms of regulated cell death pathways including necroptosis. Dr. Cao and Dr. Dovey’s expertise and research interests will be combined synergistically in the effort to understand cell death mechanisms that are critical for human health.
Publications:
A Genome-wide Haploid Genetic Screen Identifies Regulators of Glutathione Abundance and Ferroptosis Sensitivity. J.Y. Cao, A. Poddar, L. Magtanong, J.H. Lumb, T.R. Mileur, M.A. Reid, C.M. Dovey, J. Wang, J.W. Locasale, E. Stone, S.P.C. Cole, J.E. Carette, S.J. Dixon. Cell Reports. 2019, 26, 1544–1556.
MLKL Requires the Inositol Phosphate Code to Execute Necroptosis. C.M. Dovey, J. Diep, B.P. Clarke, A.T. Hale, D.E. McNamara, H. Guo, N.W. Brown Jr, J.Y. Cao, C.R. Grace, P.J. Gough, J. Bertin, S.J. Dixon, D. Fiedler, E.S. Mocarski, W.J. Kaiser, T. Moldoveaunu, J.D. York, J.E. Carette. Molecular Cell. 2018, 70, 936–948.
Lanthanide Resonance Energy Transfer (LRET)-Based Imaging of GPR126 Signaling In Vivo
Postdoctoral Scholars:
Dr. Paulina Ciepla, Chen Lab
Dr. Ana Meireles de Sousa, Talbot Lab
Mentors:
Prof. James Chen, Chemical and Systems Biology & Developmental Biology
Prof. William Talbot, Developmental Biology
Project Summary:
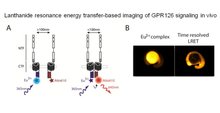
G-protein coupled receptors (GPCR) are essential for many aspects of human development, physiology, and behavior. These receptors are also important drug targets. There is intense interest in understanding how different signals activate GPCRs, but progress has been hindered by a lack of methods to visualize active receptors in living organisms. The goal of this project is to establish a new imaging method using lanthanides (metallic chemical elements) to visualize the interactions of GPCRs and other proteins in cells, tissues, and embryos. The aims are to develop an imaging approach with much greater sensitivity than current methods and to use it to visualize interactions of a particular receptor, GPR126. GPR126 controls the formation of the myelin sheath in nerves of humans and other vertebrates. GPR126 role was first discovered in zebrafish, a vertebrate model organism with fundamental similarities to humans. In addition, the zebrafish embryo is transparent, making it ideally suited for this application. Understanding the mechanisms that activate GPR126 may lead to new ideas for nerve disease therapies. Additionally, development of lanthanide-based imaging as a general approach to study a wide range of molecular interaction in living organisms would have a broad impact in many fields of the biomedical sciences.
Dr. Ciepla is a postdoctoral fellow in Chemical and Systems Biology and an expert in designing and optimizing chemical probes of protein function. During her PhD at Imperial College London (UK), Paulina’s research focused on the development of imaging tools to study posttranslational modifications of Hedgehog protein. Realizing that zebrafish provide a powerful tool in which to study the functional effects of drugs and chemical probes, she chose to further develop her skills in the lab of Dr. James Chen. Dr. Sousa is a postdoctoral fellow in Developmental Biology, with an excellent track record in biochemistry, cell biology and animal genetics. During her PhD at the Wellcome Trust Centre for Cell Biology (Scotland/Portugal), she studied the mechanisms that control microtubule dynamics and spindle formation during cell division through the combined use of forward and reverse genetics and varied biochemical approaches. During her time at in the Talbot Lab, Ana has focused on nervous system development and neuroimmune interactions. Dr. Sousa’s knowledge of GPR126 biology and zebrafish genetics together with Dr. Ciepla’s background in chemical probe-based imaging provides a unique combination of expertise to enable this important mechanistic study.
